Research
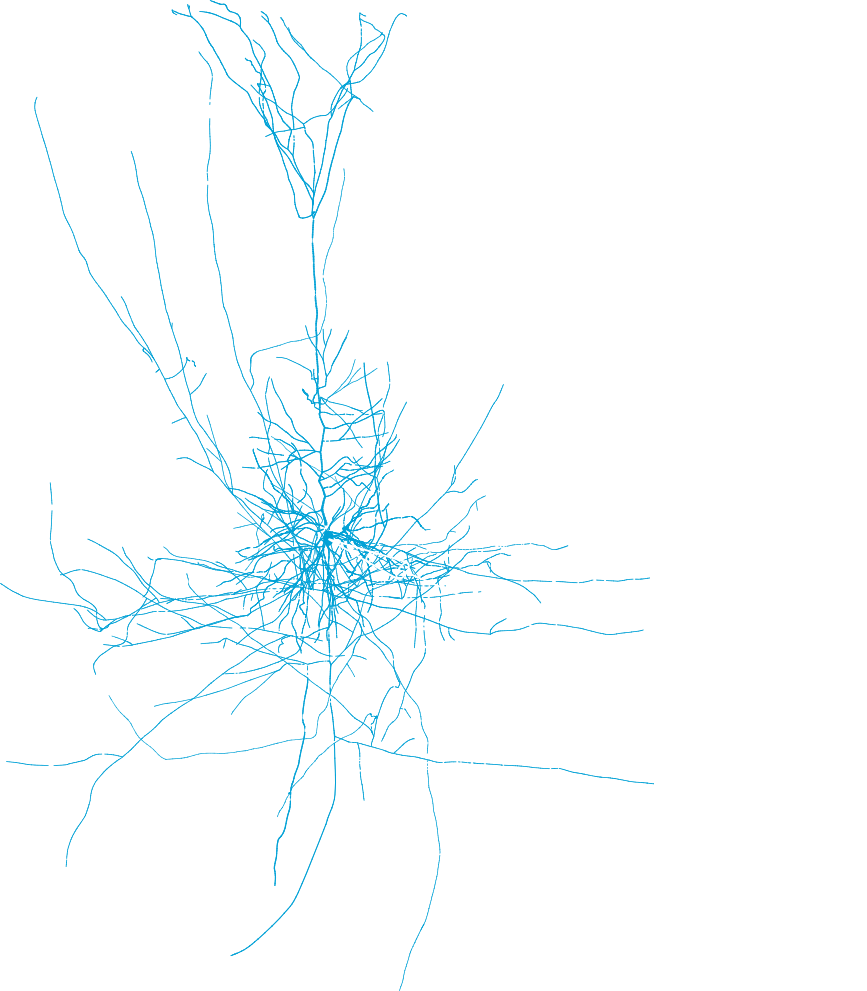
Studying the biophysical properties of neurons, such as the different ion channel distributions and kinetics, can be done by fitting in silico neuronal models to in vitro neuronal recordings. In this process, a detailed neuron is modeled to a set of electrical circuits that describe its biophysical properties. The goal is to constrain the in-silico model’s parameters so it will behave like the in vitro neurons recorded during the experiment. This process is done using an optimization algorithm that relies on training the model across thousands of permutations. We have developed a pipeline to fit the models to experimental data and we are now working on improving these models to develop highly detailed biophysical simulations for recorded neurons.
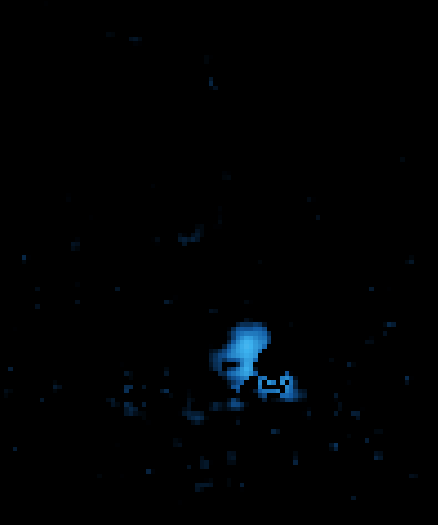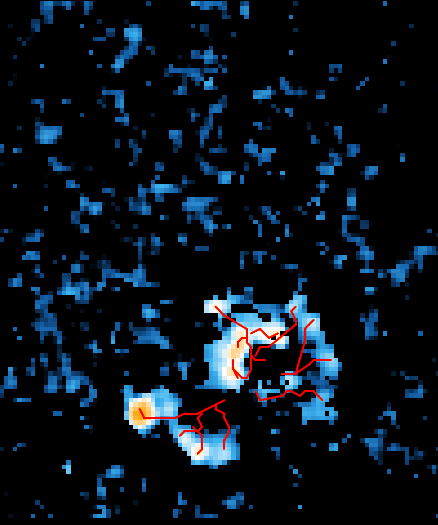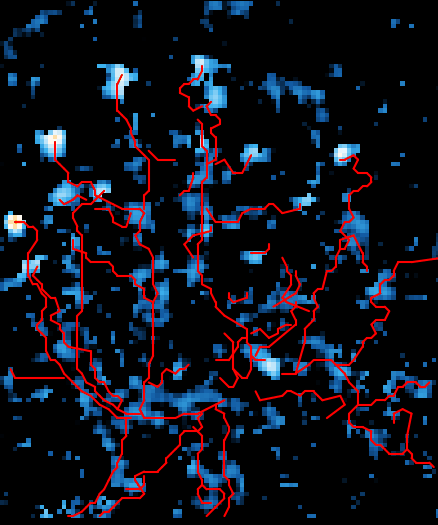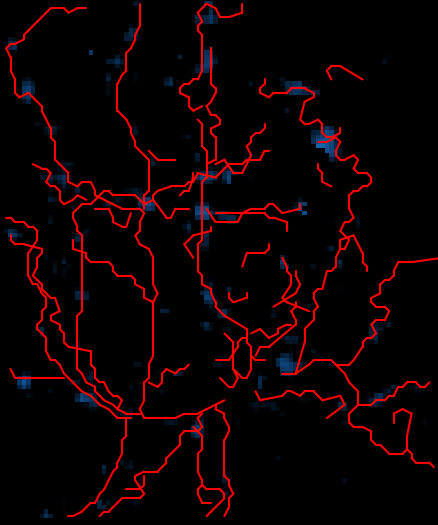
Using high density microelectrode arrays (HD-MEA), we aim to elucidate the complex neurophysiological mechanisms of neurodevelopmental conditions. This initiative meticulously analyzes the electrophysiological properties of cells derived from mouse primary neurons, human induced pluripotent stem cells (iPSCs), and organoids. This approach facilitates a comprehensive characterization of different NDDs, allowing for the identification of distinctive patterns and mechanisms at the cellular level that contribute to the manifestation of of each unique condition. 22q11.2 deletion syndrome (DiGeorge syndrome), a rare genetic condition caused by the deletion of a small segment of chromosome 22, serves as a primary focus of our research. We're using human patient-derived and CRISPR-engineered iPSC lines to study how these mutations alter neuronal network activity and exploring potential therapeutic approaches through genetic and pharmacological modulation, working closely with collaborators across disciplines. Through the integration of advanced electrophysiological techniques and diverse cellular models, we aim to unravel the intricacies of neuronal circuit dysfunctions, offering potential pathways for the development of targeted therapeutic interventions.
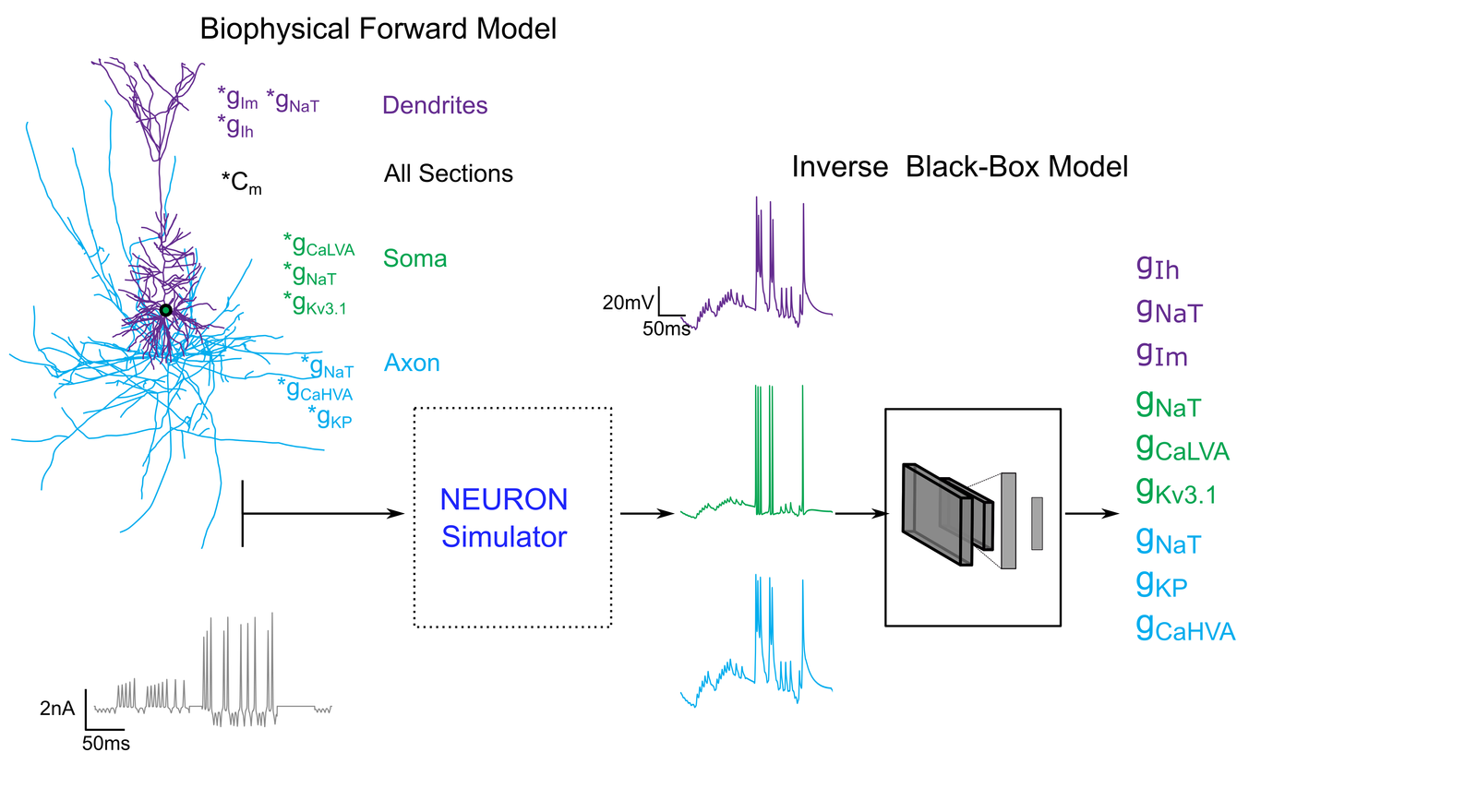
A neuron's function is defined by its unique expression of ion channels, but these are nearly impossible to measure directly. Our project solves this inverse problem by creating a Neuro-Inverter: a deep learning model trained to work backward from a neuron's electrical "fingerprint" to its underlying molecular recipe. Our model, a powerful Convolutional Neural Network (CNN) called ONTRA, learned this relationship from a massive dataset of millions of simulated neurons where the ground truth was known. Our ultimate goal is to apply this Neuro-Inverter to real-world experimental data from patch-clamp recordings. This will provide a powerful new tool to accelerate neuroscience research, enabling the rapid classification of neurons and offering new insights into channel-related diseases (channelopathies) like epilepsy and autism.
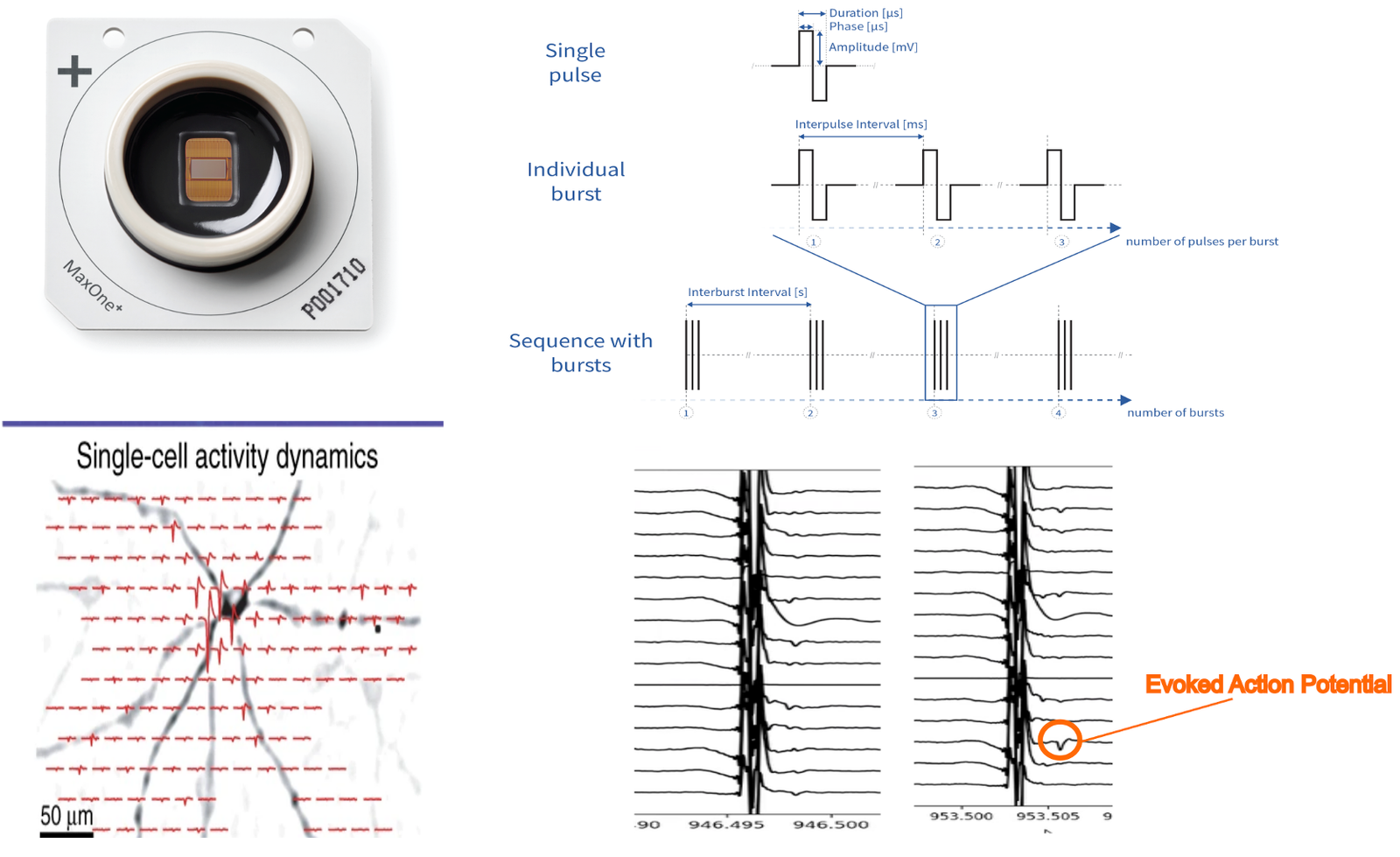
Electrical stimulation with high-density microelectrode arrays (HD-MEAs) allows us to precisely activate neurons at single-cell and network levels. By delivering controlled electrical pulses or bursts, we can reliably evoke action potentials and adjust the firing rate of neurons. This approach enables systematic study of how neuronal circuits respond to external inputs. Ultimately, it provides a powerful tool to probe excitability, connectivity, and plasticity in brain-derived cultures.
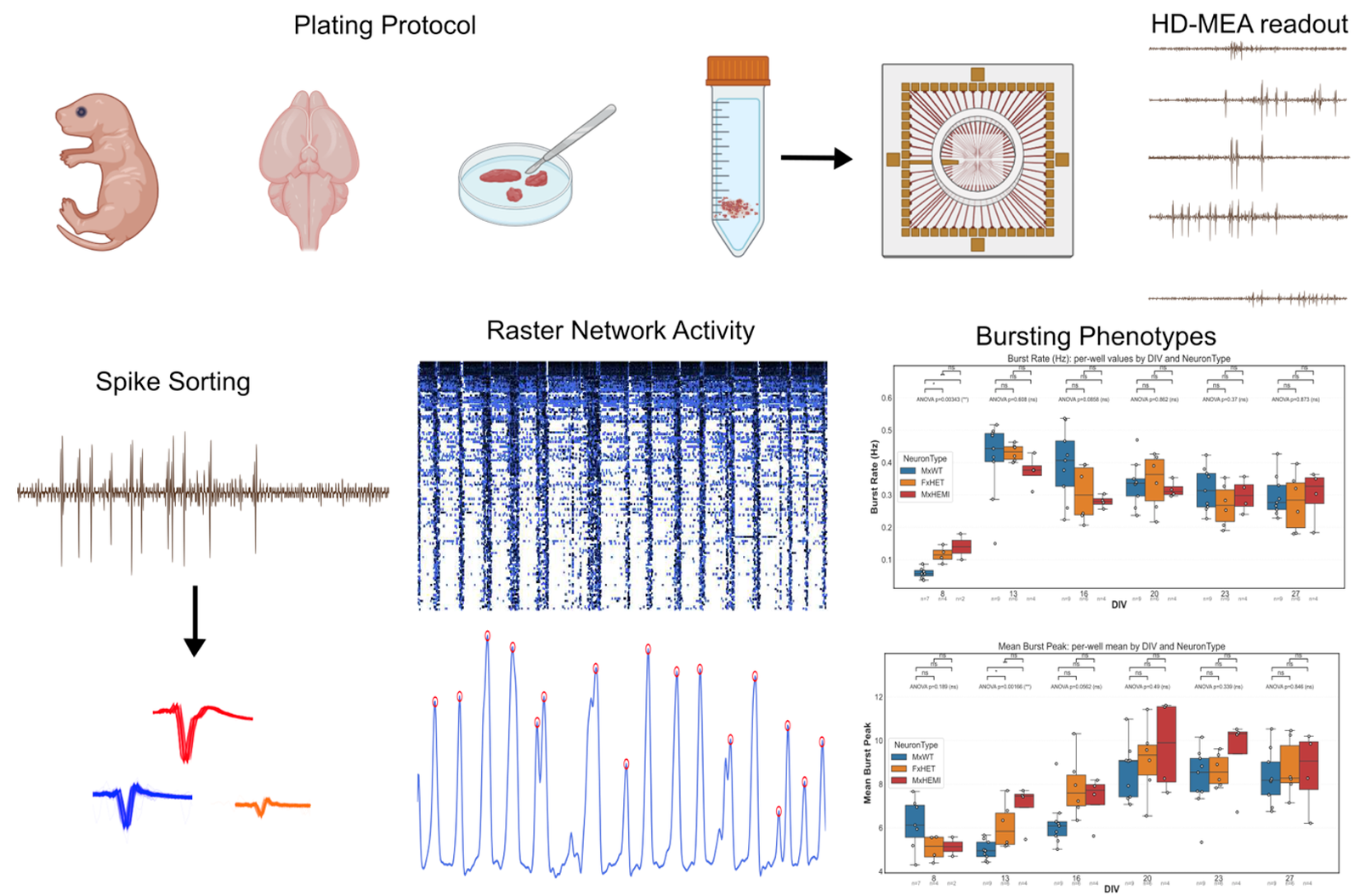
The KCNT1 gene encodes a sodium-activated potassium channel involved in regulating neuronal excitability. Pathogenic variants cause two autosomal dominant conditions: autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE) and a severe developmental and epileptic encephalopathy (DEE). Clinical outcomes vary widely and no targeted treatments currently exist. Our research aims to use high- density microelectrode array (HD-MEA) chips to record activity from wild-type and KCNT1 mutant mice frontal cortical regions. The MEA is an in vitro system that preserves native circuit architecture, allowing us to characterize electrophysiological and phenotypic properties of KCNT1 neuronal cultures and acute slices, further understanding neuronal network dynamics of genetic variants, which will help us test new pharmacological treatments to regulate its activity. Our pipeline includes spike sorting using python packages such as Spike Interface, Kilosort, and Phy, as well as our own computationally developed methods.
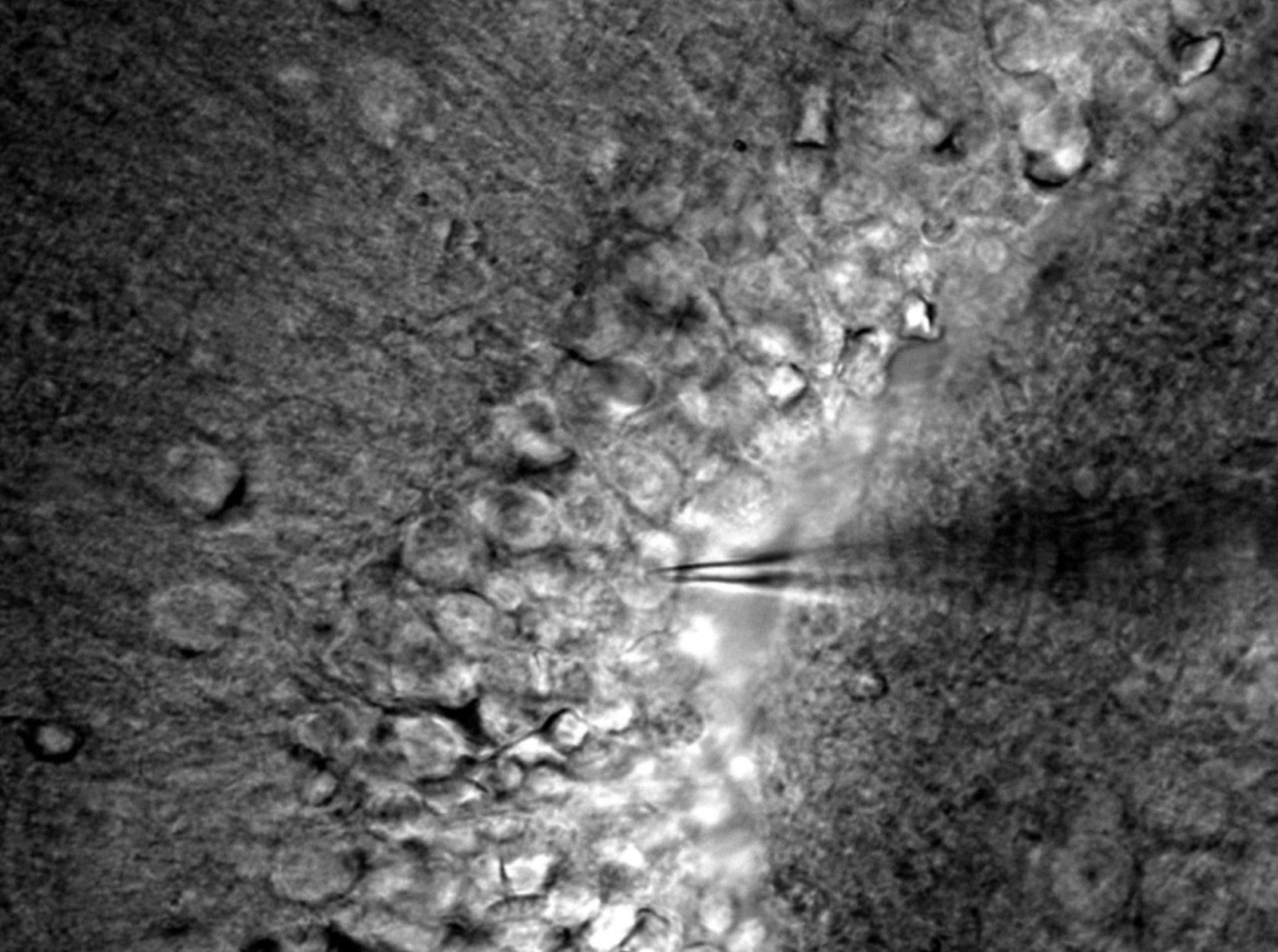
This project investigates the role of STAT3 signaling in epileptogenesis using the IHKA mouse model of temporal lobe epilepsy. We employ conditional STAT3 knockout mice to test the hypothesis that STAT3 activation is necessary for the development of neuronal and network hyperexcitability. We combine whole-cell patch-clamp electrophysiology in dentate granule cells with high-density CMOS-MEA recordings to quantify changes in intrinsic properties, synaptic transmission, and network-level synchrony and oscillations. These empirical data are integrated into Hodgkin-Huxley-based computational models to formally link the cellular consequences of STAT3 deletion to the suppression of aberrant network activity, thereby identifying and validating novel therapeutic targets.